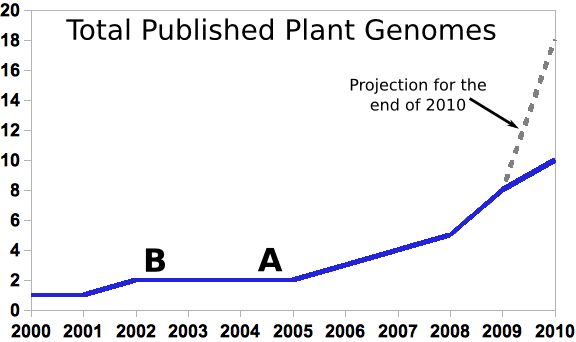
Grad students graduating with PhDs right now probably entered grad school at point A, or earlier. People at the end of their first post-doc (and potentially in a position to apply for faculty positions and start training new graduate students themselves) would have entered grad school at point B or earlier.
Today there are a mere 10 published plant genomes out of the more than quarter million named plant species in the world. But even ten genomes is a huge amount of data to deal with for a plant genomics community that largely came of age during the long genome drought of 2002-2006. What is the genome drought? The rice genome was published in April 2002. It was only the second plant genome to be sequenced, and the last plant genome to be published until the poplar genome came out in September of 2006, a gap of more than four years. Two genomes, especially of species as distantly related as arabidopsis and rice doesn’t make for a lot of compelling comparative genomics (Although there was certainly some really cool stuff being discovered in this time period.)
Does that matter? Probably not, but it’s important to remember that the people earning their PhDs today probably entered grad school (and chose a lab and field of study) during that two-plant-genome era (see point A) and less opportunity for exciting research mean less grant money and less ability to attract grad students. The youngest people applying for faculty positions today (assuming they only did one, quite successful, post-doc), also entered grad school in the two genome era (see point B), if not the previous single genome era.
I’m talking about this mostly to make the point that I think comparative genomics as a field of study is getting a lot more exciting as more genomes become avaliable, which is likely to attract more graduate students in that key first year when they join a lab and begin to specialize. Which means as we move farther away from the time of the long genome drought, we will hopefully* start to see a lot more well trained people doing plant genomics.
Which is a good thing because the other point this graph should make (not that I think many people need to be reminded of it), is that the pace of sequencing plant genomes is accelerating, and SOMEONE needs to analyze the huge quantities of data that are already starting to flow through the plant biology community.
This should be the last plant genome themed post for a while, but please continue to let me know if you know/hear about more plant genome projects. jcs98 (@) jamesandthegiantcorn.com
*If the hypothetical end to the also-hypothetical-and-possibly-the-result of-wishful-thinking-on-my-part shortage of plant comparative genomicists could hold off long enough for me to be really in demand when/if I finish grad school, that would be great. 😉
If you know how to do clever stuff with genomics data, you’ll be in demand for decades. However, anyone who does derivative stuff should be terrified of the Bejing genome sequencing project…
Comment by Matt — March 15, 2010 @ 6:11 pm
Yeah. As near as I can piece it together there was a whole separate cucumber genome project in the US that was scooped by the version published by BGI-Shenzhen. As China continues to sink more money and man hours into genomics I expect a lot more projects will end in the same disappointing way.
Comment by James — March 15, 2010 @ 9:20 pm