Hunting for good candidate genes is something biologists spend a lot of their time doing. Here are a couple of hypothetical examples:
A) Suzzy the grad student is mapping a recessive mutant which makes the pollen of cornplants shrivel up and die. By examining a bunch of known genetic markers in plants with dead pollen and normal pollen producing siblings of those plants she has narrowed the location of the gene responsible for her trait down to a region of only a couple of megabases on the fifth chromosome of maize. Since the whole maize genome contains over 2,300 megabases of sequence that means she’s already ruled out 99.9% of the genome. But her region still contains, say, a dozen genes and she needs to know which one she should check first to see if mutation in it is responsible for her mutant phenotype.
B) Johnny is another grad student. He wants to understand how corn plants genetically regulate how wide their leaves will grow to be. By measuring a lot of plants descended from two parents, each with known genotypes, he can identify regions of the genome where inheriting information from one parent or the other seems to be correlated with either wider or narrower leaves. He calls these regions quantitative trait loci (or QTLs). Now he has picked the genetic region that seems to have the biggest effect, and he wants to know what gene within the region is actually responsible for the effect.
There are a number of ways for both Johnny and Suzzy to narrow down their lists to the genes most likely responsible for the changes they are each observing in corn plants:
- A okay candidate might contain a protein domain with a function related to the plant trait they were studying.
- A good candidate might show a specific pattern expression in the part of the plant where they observe their phenotype (pollen for Suzzy and developing leaves for Johnny).
- A GREAT candidate would be a gene which has already been studied by another group and has a mutant phenotype related to the phenotype currently observed.*
Checking out the protein domains of genes under a QTL or mutant interval can be accomplished in almost any genome browser (for example the ones provided by MaizeGDB, or CoGe).
Checking the expression patterns of genes is harder. One option for plants is a website called PlexDB which lets you look up the expression of individual genes in different people’s microarray experiments. The biggest problem with microarrays is that it really is impossible to compare expression between different people’s microarray experiments.
Identifying known mutant genes within a genomic interval used to be a real pain. Now they could search the classical maize gene list to see if any of the genes in their interval appear on it, but it is still not what you’d call efficient.
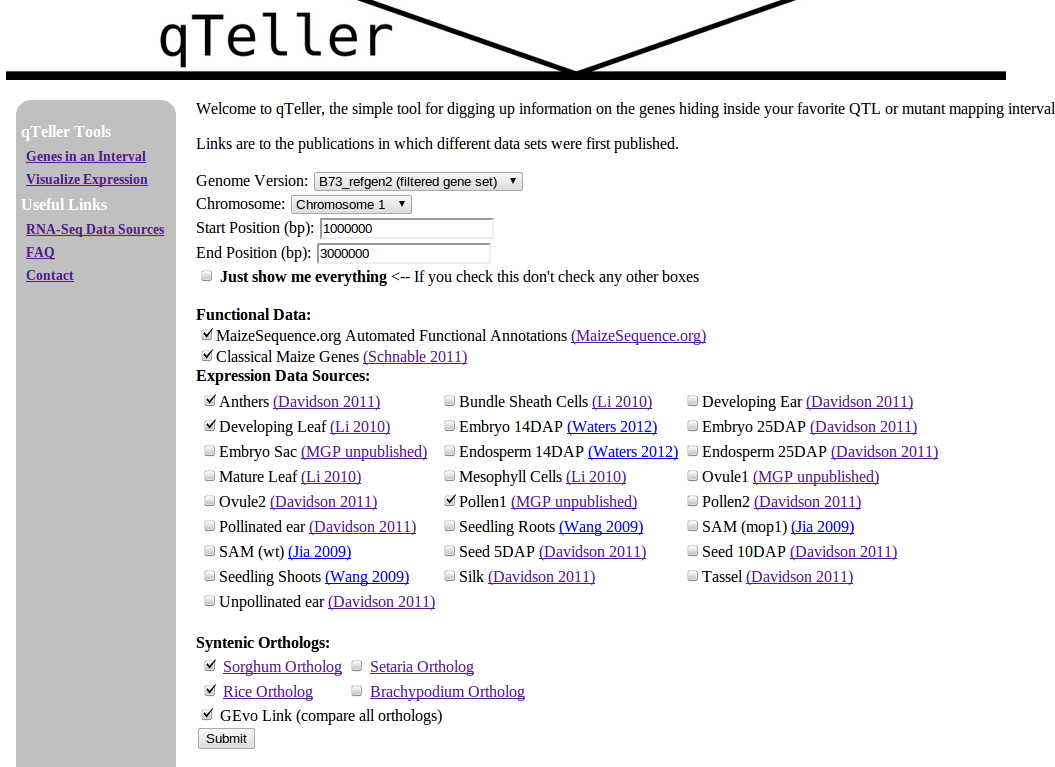
Now imagine a website that let you check all those things in one step!
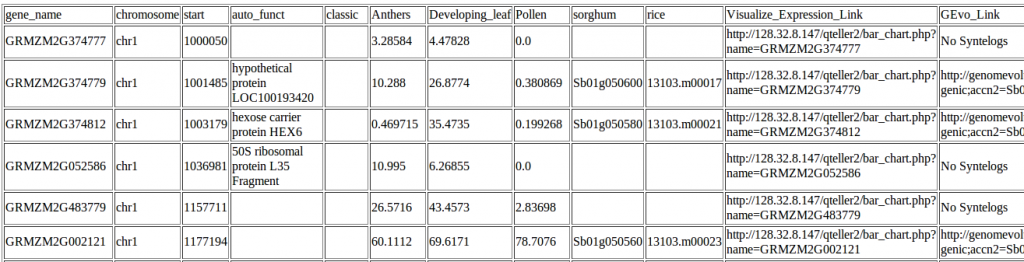
Example of data on genes in a region 10 megabases from the start of maize chromosome1. Click to zoom in and make the text big enough to read.
The automated annotations of protein domains are the same ones you’d find in any of the genome browsers above, but instead of checking each gene in turn, qTeller reports them all in a handy sortable spreadsheet (which you can either view online or download to your computer).
It also incorporates measurements of gene expression using RNA-seq data from papers published by the maize community.** To make the numbers as comparable as possible I went back to the raw reads provided by NCBI’s Sequence Read Archive and taken each dataset through the same analytical pipeline.
And, of course, it reports any classical maize genes which lie within the interval a researcher is studying (taken from the version two classical maize gene list.)
Plus syntenic orthologs in other species. Because, at least in the grasses, genes with mutant phenotypes are disproportionately likely to have been retained at the same location in the genome of lots of different grass species.
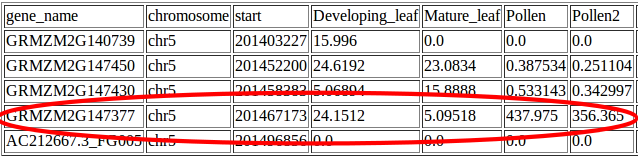
Suzie found a gene within her interval which was expressed at much higher levels in pollen than in leaf tissue:
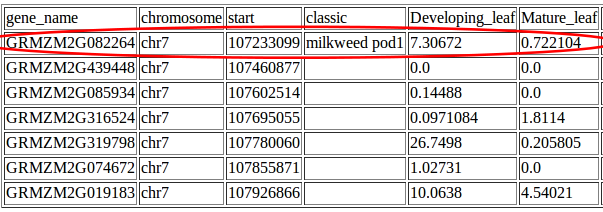
And Johnny realized that his QTL contained the mutant gene milkweed pod1, a classical maize mutant known from previously published papers to be involved regulating in leaf development.
So yeah, qTeller is what I’ve been working on for the past few months. Please let me know if you run into any bugs or have any questions. -James
*Admittedly this would be rather disappointing news for the grad students involved (it’s a lot harder to get a splashy paper out of rediscovering a known mutant), but it’s much better to find out you might be studying a known gene early so you can check and cut your losses if it turns out to be true, instead of after you sink another couple of years into recloning and recharacterizing it.
**The authors of all these papers deserve a whole bunch of credit for generating these datasets:
- Waters AJ, Makarevitch I, Eichten SR, Swanson-Wagner RA, Yeh C-T, et al. (2011) Parent-of-Origin Effects on Gene Expression and DNA Methylation in the Maize Endosperm. The Plant Cell doi:10.1105/tpc.111.092668.
- Davidson RM, Hansey CN, Gowda M, Childs KL, Lin H, et. al. (2011) Utility of RNA Sequencing for Analysis of Maize Reproductive Transcriptomes. The Plant Genome4:191-203 doi:10.3835/plantgenome2011.05.0015
- Li, P., Ponnala, L., Gandotra, N., Wang, L., Si, Y., et al. (2010) The developmental dynamics of the maize leaf transcriptome. Nature Genetics 42: 1060-1067. doi:10.1038/ng.703
- Wang, X., Elling, A.A., Li, X., Li, N., Peng, Z., et al. (2009) Genome-Wide and Organ-Specific Landscapes of Epigenetic Modifications and Their Relationships to mRNA and Small RNA Transcriptomes in Maize. Plant Cell 21: 1053-1069. doi:10.1105/tpc.109.065714
- Jia, Y., Lisch, D.R., Ohtsu, K., Scanlon, M.J., Nettleton, D., et al. (2009) Loss of RNA Dependent RNA Polymerase 2 (RDR2) Function Causes Widespread and Unexpected Changes in the Expression of Transposons, Genes, and 24-nt Small RNAs. PLoS Genet 5: e1000737. doi:10.1371/journal.pgen.1000737
- The Maize Gametophyte Project: Unpublished Dataset SRP006965
Hi.
Just wanna to say that your blog and work were much appreciated on our lab yesterday.
Hope you keep up the good work. 😀
best of luck.
Comment by Ana Catarina — May 5, 2012 @ 4:02 pm
Thanks Ana!
Comment by James — May 5, 2012 @ 4:07 pm
Its very informative tool. Please let me know how to cite this tool.
Comment by Bhavani — August 31, 2012 @ 4:32 pm