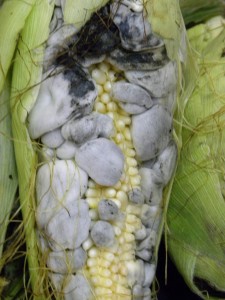
And no that doesn’t mean corn pornography*. Corn smut, or Ustilago maydis, is a fungus that infects corn plants. It’s an old acquantance from my days working in the field. We always used to tell the new hires that corn smut was a rare delicacy in some countries (as we’d been told ourselves), but this was in the days before iPhones so until recently I never actually checked on this bit of received wisdom.
Turns out this particular bit of knowledge was true:
The immature galls, gathered two to three weeks after an ear of corn is infected, still retain moisture and, when cooked, have a flavor described as mushroom-like, sweet, savory, woody, and earthy.
I haven’t been able to figure out what the trade off in nutrition is between the ear of corn that is produced by a normal plant and the fungal galls that can be harvested from a plant infected with corn smut. I’d imagine corn smut provides more (and more complete) protein than an ear of corn (assuming corn smut is nutritionally similar to mushrooms.) But what’s the comparison in number of calories? The fungus is certainly sold at a higher price pound for pound.
My renewed interest in corn smut comes courtesy of a new paper** that came out in PLoS Biology describing how the fungus steals energy from infected corn plants without triggering the corn’s usual anti-fungal defenses. It’s an interesting read, you can check out the paper itself since PLoS Biology is open access, or Diane Kelley’s summary at “Science Made Cool.”
I’d seen a number of talks recently about another fungal parasite, powdery mildew in Arabidopsis, but somehow it’s much easier to focus on this stuff now that I can connect it back to corn. Even mammalian systems can be interesting*** once the make that connection.
*Please PLEASE don’t let that phrase start showing up in the search terms people use to find my site!
**Wahl R, Wippel K, Goos S, Kämper J, Sauer N (2010) A Novel High-Affinity Sucrose Transporter Is Required for Virulence of the Plant Pathogen Ustilago maydis. PLoS Biol 8(2): e1000303. doi:10.1371/journal.pbio.1000303
***The talk I’m practicing for Monday actually uses an example of a pheromone receptor in new world monkeys that was lost 23 million years ago in old world monkeys (including us humans).