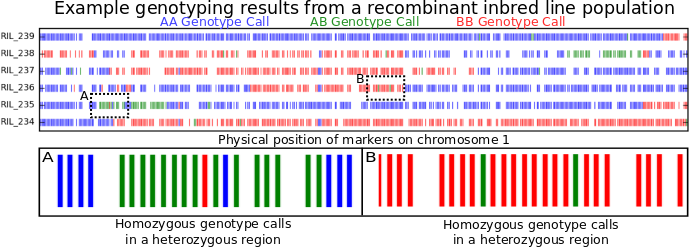
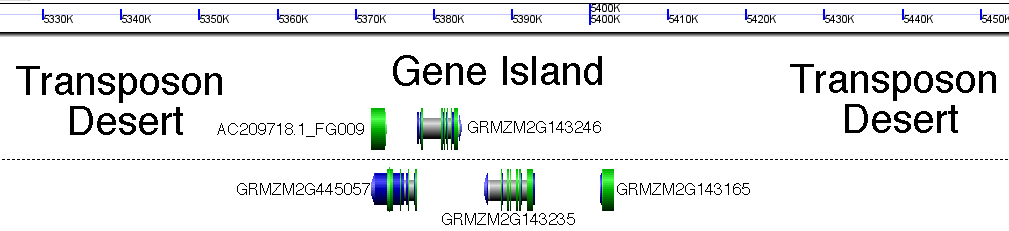
I’m just about wrapped up with the big project I’ve been working on recently. Hope to be able to say more about it in the not-too-distant future. Having to be secretive in science sucks.
But there’s a lot of be happy about! I’m done teaching for a long time. As much as I enjoyed working with the kids in my class, the other responsibilities of teaching (grading, sitting through lectures without the chance to break in for the discussions and arguments that make academia so fun, grading, designing assignments, grading) were really starting to wear me down.
And I’m only three weeks (June 22nd) from either passing my qualifying exam or becoming a beaten and broken shell of a man. For three hours four professors will question me on everything I’ve learned (or should have learned but didn’t) in my education up to this point, and everything I propose to spend the next few years of my life doing. This may not sound like a good thing, but it is. Because my qualifying exam has been hanging over my head all semester,
The lab has a new paper in press, having run the sequential gauntlets of Peer Review, Editorial Evaluation, and finally (and perhaps most dreaded) Your-Figures-Aren’t-High-Resolution-Enough e-mails from the journal’s publication department. But more on the details of that whenever the paper actually shows up.
But what was the point of this entry again? Oh yeah. Transposons. I have a soft spot from transposons (I’m guessing most people who work with maize genetics do). Today we may know that transposons are found in practically every genome under the sun, but they were discovered first in maize using old school genetics (breeding plants together and counting traits in the offspring), before DNA sequencing was a gleam in its inventor’s eye.
And on top of that, some delightfully high-copy number transposons are in the middle of proving a major scientific point for me, so I figured the least I could do was devote a week to them here on the site.
If you’re not a geneticist, should you still care about transposons? Absolutely! Transposons are one of the best arguments, not for why genetic engineering is safe, but for why, if anyone worried about hypothetical unintended consequences of genetic engineering should be worried about any food with DNA in it (and as far as I know, that’s all food.) To paraphrase a seinfield character: “No food for you!”
The week’s schedule: (more…)