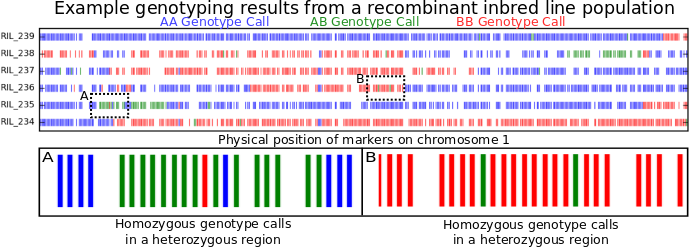
When doing anything even vaguely related to quantitative genetics I would chose more missing data over more genotyping errors any day of the week. There are lots of approaches to making missing data less of a pain. The most straightforward of these is called imputation. Imputation essentially means using the genetic markers where you do have information to guess what the most likely genotypes would be at the markers where you don’t have any direct information on what the genotype is. This is possible because of a phenomenon known as linkage disequilibrium or “LD.” Both imputation and LD deserve their own entire write ups and they are on the list of potential topics for when I have another slow Sunday afternoon. For now the only thing you have to know about them is that, when information on a specific genetic marker is missing, it is often possible to guess with fairly high accuracy what that missing information SHOULD be. But when the information on a specific genetic marker is WRONG… well it’s usually a bit more of a mess (but I think the software solutions for this are getting better! Details at the end of the post.)