A group of researchers at Masaryk University in the Czech Republic study how different members of the same grass species (Festuca pallens) have different total amounts of DNA per cell. have a new paper out in New Phytologist they found that plants with the most unusual genome sizes (really big or really small) are less likely to survive to adulthood than plants that have roughly average amounts of DNA per cell. In other words, plants with abnormally high or low genome sizes have lower fitness than their averagely genome endowed siblings.
One point the authors don’t get into, is whether what they’re really seeing is evidence of heterosis (hybrid vigor). Which is not to say they haven’t thought of it. I can’t think of any way to address the question in a non-model system like Festuca, so any mention of it would have been the sort of pure speculation some peer reviewers don’t care for. But I, writing a totally un-peer reviewed blog, am free to speculate to my heart’s content:
I’ve talked before about how plants, like people, contain two copies of their entire genome, one from their father, and one from their mother. That means any given part of the genome has the potential to be present as two dissimilar copies. Now imagine that for most locations in the Festuca genome, one of those versions is bigger than the other, sometimes by a little, sometimes by a lot.* The plants with the smallest genomes in the next generation will be the ones that inherited two copies of the smaller versions of many pieces of the genome, and the plants with the biggest genome will have inherited two copies of the larger versions of many pieces of the genome. That’s a complicated idea, so consider this simplified example:
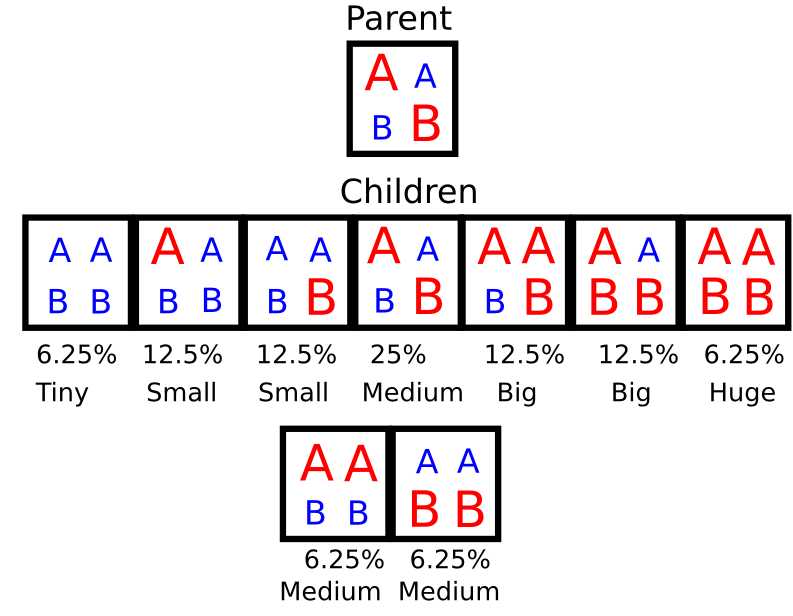
A and B represent two different parts of a genome. Think of them as genes, chromosomes, whatever works for you. The larger version red letters present versions that contain more total DNA, and the smaller, blue letters represent shorter equivalent versions of the same part of the genome. The percent of children that will display each arrangement, and the size of genomes with that arrangement is marked below. *Thanks to Sanjuro for pointing out an oversight in an earlier version of this graphic.
Why would that matter? Because having diverse versions of genomes results in more vigorous healthy plants (known as hybrid vigor or heterosis), and having less diversity in your genome results in sickly, unhealthy plants (called inbreeding or inbreeding depression). So if plants with extreme genome sizes tend to have less genetic, it would make sense to expect those plants to be less likely to out compete their more vigorous neighbors long enough to survive to adulthood (especially since, as the authors point out, only one in 15,000 seeds survives long enough to reproduce on its own).
Wouldn’t it?**
Another explanation, which the authors of the paper put forward, is that the largest and smallest genome sizes may result from chromosomal rearrangements where significant portions of the genome are lost or duplicated, which would obviously cause major problems for the resulting plants. Whatever the real answer is, discovering it will require further study.
I only found out about this paper because I have a filter to grab any new research articles that mention tetraploidy (whole genome duplication) and by pure luck Festuca pallens is a tetraploid grass.***
I’m hoping someone will do a similar experiment in corn. Different lines of corn have significantly different amounts of DNA per cell (a quick search of the literature turned up a reference claiming variation of up to 40%) and maize community has great sets of markers to assess how much genetic diversity is preserved in the genomes of different plants. The downside of course is that, as a domesticated crop, “fitness” is a more poorly defined concept for corn than it is for a wild species like Festuca pallens.
*Why is it bigger? Maybe there are more transposons (my review of what transposons are) in one copy than the other. Maybe genes have been lost from one version but not the other (hey it happens in maize!).
** I’m certainly not an ecologist****, so there’s a real risk I’m missing something obviously wrong with this idea.
***Is it luck? Do tetraploid species have larger than average variation in total genome size? I could see any argument for why it might be so, but I don’t know if anyone has checked.
****I’m also not a population geneticist. I’m not a quantitative geneticist. I’m not a biochemist. I’m not a botanist. I’m certainly not a real computer programmer. And those are the specialties I’ve wished I was an expert on just the past week!
Šmarda, P., Horová, L., Bureš, P., Hralová, I., & Marková, M. (2010). Stabilizing selection on genome size in a population of Festuca pallens under conditions of intensive intraspecific competition New Phytologist, 187 (4), 1195-1204 DOI: 10.1111/j.1469-8137.2010.03335.x

I know this wasn’t really the point of your post, but shouldn’t there be a couple more genotypes in the image (aaBB and AAbb)?
Interesting post though, as usual 🙂
s
Comment by Sanjuro — August 24, 2010 @ 1:14 pm
Hahaha! Yes, you’re right. I guess it’s been too long since I drew out a real two trait Punnett square. Updated graphic forthcoming….
Comment by James — August 24, 2010 @ 2:24 pm
Yeah, I had to draw one out to convince myself 🙂
Comment by Sanjuro — August 24, 2010 @ 2:49 pm