Out of the ~12,000 known grass species, the genomes of less than one in one thousand have been sequenced. The “One in a Thousand” series focuses on these rare grass species.
Dichanthelium oligosanthes is a wild grass that grows in forest glades throughout the American midwest. It is a small plant. Doesn’t grow particularly fast. Its flowers aren’t particularly striking. And it has enough issues with seed dormancy that growing it in captivity is a major pain. Dichanthelium is a one in one-thousand grass with a sequenced reference genome.*
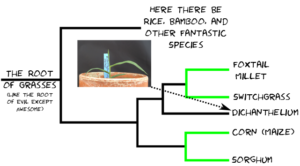
The reason folks are interested in Dichanthelium isn’t because of what it is, but who it’s related to. Dichanthelium occupies a spot on the grass family tree between a tribe** of grasses that includes foxtail millet and switchgrass, each one in a thousand species themselves, and another tribe of grasses that includes corn and sorghum, two more one in a thousand species. The relationship looks something like this:
The species highlighted in green use C4 photosynthesis.*** Photosynthesis has been around for a LOOOONNNNNGGGG time. It’s an extremely optimized process. Think of photosynthesis as the internal combustion engine. There were a lot of efficiency gains in the early years, going from a Ford Model T to a Ford Model A and so forth. But now, after decades of optimization, it is harder and harder to build an internal combustion engine that makes major steps forward in efficiency. Then someone comes up with the idea of strapping an electric motor (an existing piece of highly optimized technology used for completely different things) into the drive train. Cars still have all the same parts they did before, but they’ve added some extra systems that let them drastically improve efficiency (although there is still a lot of optimizing to be done).
The difference between conventional photosynthesis and C4 photosynthesis is like the difference between an internal combustion engine and a hybrid electric drive train. In both cases the latter adds a bunch of additional equipment to what is already there, making things more complex, but also potentially more efficient. In the case of plants, C4 photosynthesis lets them capture carbon dioxide and turn it into sugar more efficiently (and using less water), letting C4 plants thrive and out compete their relatives, particularly in hot and dry environments.

A younger Dichanthelium plant that’s just starting to flower (it flowers twice a year once in the spring and once in the fall). Honestly they’re kind of cute if you can forgive the poor germination.
Now imagine you knew absolutely nothing about cars, and someone asked you to make a list of all the parts that were unique to hybrid electric cars orwhich had significantly changed in such cars. The naive approach (which has a direct analogy to what we do in genomics) would be to take apart a hybrid electric car and a conventional internal combustion engine car, make a list of all the parts in each, subtract out all the parts in the internal combustion engineer car, and whatever leftover parts you had from the hybrid electric car (giant lithium batteries, electric motors, regenerative breaks, etc) are the parts that make it a hybrid electric car.
Now someone gave you this job, one of the things you’d definitely want to do is pick two cars that were as closely related as possible, EXCEPT for the difference in the type of engine they used. Comparing the parts that make a silverado and those that make a prius isn’t going to tell you much. But until recently, that’s pretty much what the people who wanted to study C4 photosynthesis is crops like foxtail millet, sorghum, and maize (corn) had to do. Because the most closely related non-C4 plant with a sequenced genome they have to compare to was rice. Sequencing dichanthelium, which a much closer relative of these C4 grain crops means many more of the differences identified will actually be things that are involved in C4 photosynthesis (like comparing a Chevy Volt and a Chevy Cruze).
That’s why we put up with its slow growth rates and annoyingly poor germination, and why Dichanthelium oligosanthes made it into the one in a thousand club.
*Studer, A.J., Schnable, J.C., Weissmann, S., Kolbe, A.R., McKain, M.R., Shao, Y., Cousins, A.B., Kellogg, E.A., and Brutnell, T.P. (2016). The draft genome of the C3 panicoid grass species Dichanthelium oligosanthes. Genome Biology 17, 223.
**Yes, grasses come in tribes (and even subtribes). It’s all Carl Linnaeus’s fault. He made all grasses a single Family, and, because King Philip Can’t Order Fried Greasy Snails, the only two official taxanomic groups left below the family level were Genus and Species. The grass family tree is a bit too complex for that to work out well so we insert three extra unofficial levels — “Subfamily”, “Tribe”,”Subtribe” — between Family and Genus just to keep track of who is related to who.
**Why is it called C4 photosynthesis? (A phrase I’ve used so many time in this post I’m likely on some sort of a watch list?) Because unlike conventional photosynthesis where the first sugar like molecule produced from carbon dioxide has three carbon atoms in it, in C4 photosynthesis, the first sugar like molecular produced has four carbon atoms in it. So no, to the best of my knowledge there’s no C1, C2, or C5 photosynthesis.