“My budget…triples the number of National Science Foundation graduate research fellowships. This program was created as part of the space race five decades ago. In the decades since, it’s remained largely the same size –- even as the numbers of students who seek these fellowships has skyrocketed. We ought to be supporting these young people who are pursuing scientific careers, not putting obstacles in their path.” – President Obama
I’m still feeling brain dead after the final push for submitting.
Speaking of NSF, here’s the one figure I managed to shoehorn into my research proposal.
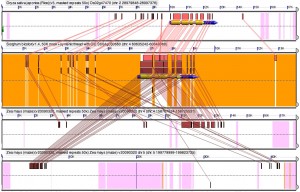
Blast hits between an orthologous quartet of gene spaces, one in rice, one in sorghum and the two copies created by the maize tetraploidy. As usual click the picture to see it fullsized
If you’d like you can even click here to be able to play around with the figure yourself using the CoGe interface. Now I’ve got to try to explain what this figure is about.
The definition of ortholog boils down to being the same gene in different species. Scientists (like me!) can identify orthologs not just by looking for the most similar gene in another species’ genome, but also looking at synteny, the order of genes on chromosomes. Orthologous genes are in the genomes of two species descended from a common ancestor*, and the genes themselves can both trace their history back a gene sitting on a chromosome in that ancestral species. After the two species went their separate ways, each of their genomes began to accumulate differences (mutations). Eventually it can be hard to tell which genes are most closely related to each other, but one of the things that lasts a long time is orthologous genes will has some or many of the same genes nearby on the chromosome, in a similar order to each other, since many genes are still in the same places in the genome as they were in the genome of the common ancestor of both the species being studied.
That’s a huge mouthful. And it gets even more complicated when plants start holding on to extra genomes (something that isn’t too uncommon in plants, see my discussion on the genetics of wheat). Maize (corn) went through such a genome doubling, estimated to have happened ~10 million years ago. So when we compared the genome of corn (which should be getting officially published really REALLY soon) to that of other grass species like rice and sorghum (all the grains we grow (wheat, rice, corn, sorghum, barley, rye, oats, etc) are grasses) there are actually two separate maize regions that are equally related to every sorghum or rice region.**
This figure shows a gene conserved between all four regions and the dna surrounding it. The lines connect blast hits, regions a program (called BLAST) used by lots of biologists as identified as having similar DNA sequences between the the segments from the different genomes.
*One of the reasons comparative genomics is a field that wouldn’t exists if the people who don’t believe in evolution were right.
**That doesn’t mean maize has two copies of every gene present in sorghum and wheat. Right after getting an extra genome copy (technically two extra copies, since normal diploid organisms have two, one from each of their parents to start with, and maize doubled both) it did. Over the past ten million years the extra copies of many genes were lost, either deleted from the genome or accumulating so many mutations that they stopped functioning.