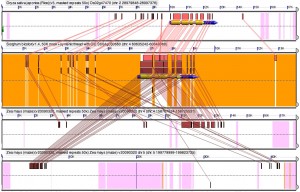
I invested in a new video capture program that lets me record voice overs in real time. There are a few more ums and uhhs but my voice and the action on screen are in much better sync, and I don’t find myself rushing to keep up with the movements of my own mouse or trying to fill apparently dead time while nothing happens on screen. The new video is also two minutes and fifteen seconds shorter, dropping below the psychologically important 5 minute barrier, above which watching a video starts to feel a lot more like work, and I get to show off two new features (visually flagged tandems, and predictions of where a gene would have been before it was lost) that we’re still in the process of rolling out.
Sorry to harp on the same topic a second time, this video is just SO MUCH BETTER than the previous one and I needed to show it off. 😉