Sorry this is late going up. -James
This morning Nature officially published the paper* describing the sequence of the Brachypodium distachyon genome. This publication brings the number of grass genomes available for comparative analysis to four. In celebration I’m going to list four reasons to be excited about the publication of this genome.
The location of Brachypodium within the grass family tree.
Brachy (as I will refer to the species from here on) is a member of the Pooideae a sub-family of grasses from which no sequenced grasses have come. For the work we do in my lab this is exciting because it adds more depth to our analysis of changes in the grass genomes. The more distantly related grasses we can compare at the whole genome level, the better we can infer what the ancestral species that gave rise to all the grasses might have been like at a genome level. The most we know, or can make educated guesses about that species, the better position we are in to say what changed along the evolutionary paths leading to grasses like maize, rice, and sorghum. The choice of the Pooideae wasn’t at random, or even because of the sub-family’s distant relationship to other sequenced grasses.
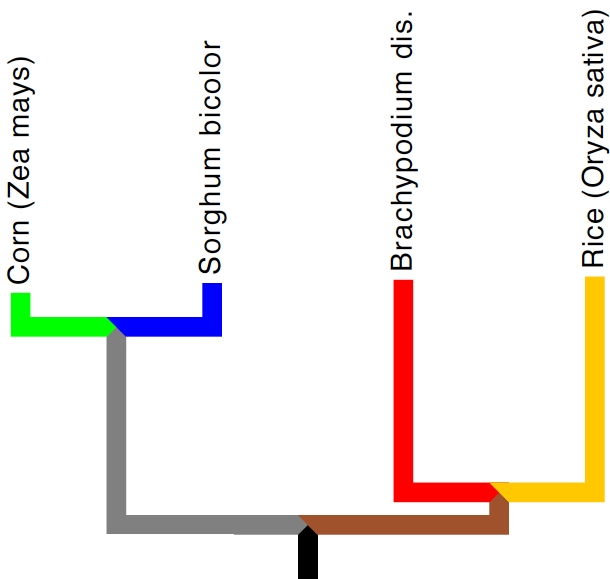
Family tree of the sequenced grasses. As you can see, corn (green) and sorghum (blue) are quite closely related, and while brachypodium (red) is more closely related to rice (orange) than it is to the sorghum/corn pair, it's still pretty far away from anything else yet sequenced. Tree visualized in Mesquite ( http://mesquiteproject.org/ ) and is an approximation at best.
As I’ve said many times, both on this site and elsewhere, it is the unrivalled productivity of three grasses (grains) that underpin our civilization. Without corn/maize, rice, and wheat, it might have been that human societies would never have been able to produce enough surplus food so that farmers could support philosophers and copper workers and all that great surplus people who do things other than bring food from the ground. Of the big three grains, rice was the second genome ever sequences and the genome of maize/corn was published this past November. Wheat stands alone as a genome so complex the very though of trying to assemble it makes grown bioinformaticians cry (I’m obviously taking some dramatic license here). As you may have guessed, wheat (and its close relatives barley, rye, and oats) also belong to the Pooideae. Prior to the publication of the brachy genome, wheat geneticists would have to go all the way to rice to find the most closely related species with a sequenced genome. So while the publication of the brachypodium genome may not be of huge excitement to wheat geneticists (the relationship between brachypodium and wheat still last shared a common ancester more than 30 million years ago), it’s still an improvement from their previous situation. (Though it may be cold comfort wheat geneticists, remember the rest of us plant folks are in awe of you.)
Brachypodium Really is the Arabidopsis of the Grass World.
It can be said as an insult, implying that like Arabidopsis, brachy is small and boring, but being small really does have its advantages for research. A lot more brachypodium plants can fit into a given square foot than can corn or sorghum. And from my own experience, brachy takes much better to life in a growth chamber than any other grass species I’ve worked with, which means instead of doing genetics out in a field, with all the costs**, limitations***, and risks**** that entails. Brachy researchers can just grow their plants in growth chambers down the hall (or downstairs) from their labs, a convenience arabidopsis researchers have been enjoying for decades. Personally I think those limitations build character and encourage the development of good habits like planning out one’s research in advance (including fallbacks and alternative avenues), but I’d also be thrilled to see more labs get into grass genetics so on the balance I consider brachy’s arabidopsis-like nature to be a Good Thing.
Brachypodium has a very NICE genome.
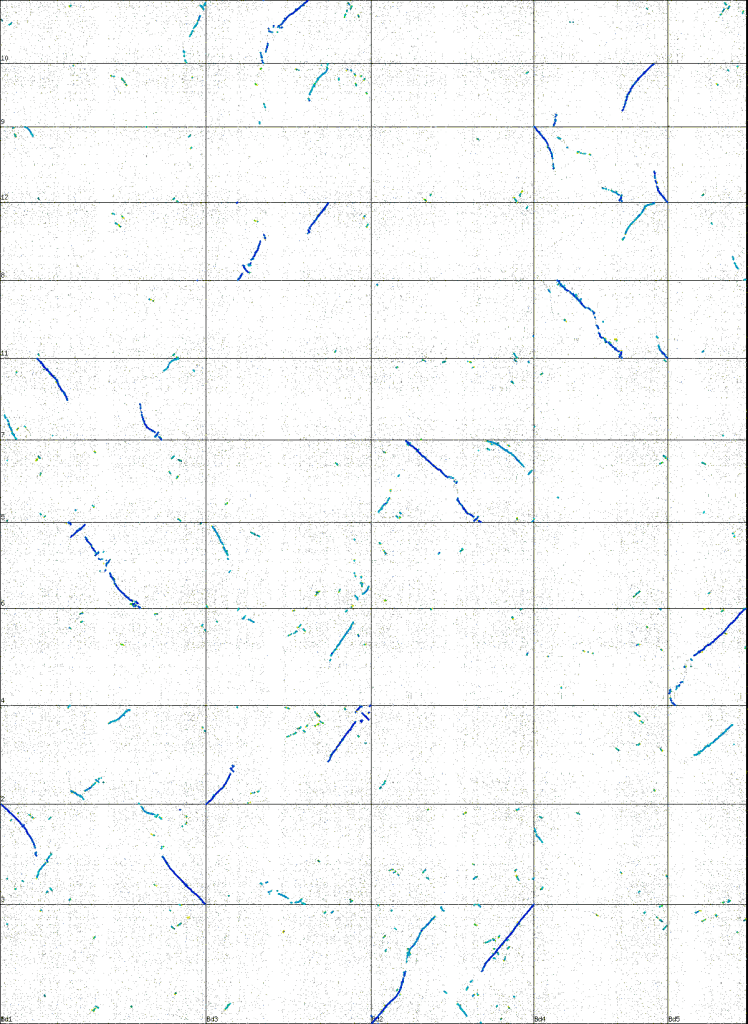
A dotplot showing the conserved order of genes in the chromosomes of rice (Oryza sativa) and Brachypodium distachyon. The darker blue diagonal lines represent orthologous genes. The light-blue/cyan lines homeologous regions (ones that were created when the ancestor of all sequenced grasses doubled its whole genome. The regions have evolved independently since but enough duplicated copies of genes are still in the same order that they're easy to spot.) Tree generated using CoGe's Synmap tool ( synteny.cnr.berkeley.edu/CoGe/SynMap.pl ), with color coding based on synonymous substitution rates.
Brachy has only five chromosomes, and, as you can see in the dotplot comparing brachy to rice, the genes line up in long straight syntenic stretches. Even the light blue regions which result from a whole genome duplication in the ancestor of all grasses are relatively long and well defined. You’ll also notice brachypodium only has five chromosomes, to rice’s 12. (Maize/corn and sorghum both have 10)
Transposons, the “jumping genes” that make working in maize so complicated, are a much smaller proportion of the total genome in Brachypodium than in other systems weighing in at less than 27% of the total genome which in total is only 271 megabases long (less than 15% the size of the 2.3 gigabase corn genome which is 85% transposons, which … mumble…carry the one …. means corn has 26.7 TIMES more transposon sequence than brachy, and I sure want to learn more about how brachy keeps its transposons in check.)
The Author List
Publishing a good genome paper is an enormous undertaking, involving collaborations between dozens of research groups across the country or sometimes around the world. Of the, by my count, 135 names attached to the paper paper I can count old employers, current collaborators, science friends and acquaintances, one relative and (at least) one regular reader of this site.
One of those 135 names is also my own. This is from work I did back during my second rotation last winter doing manual verification and analysis of flowering time genes in the brachypodium genome sequence. A very small part of the work that in turn went into a small section of the paper, but this is the first time my name has actually been attached to a peer reviewed publication, EVER! (My undergraduate work made it onto a couple of poster abstracts but no papers, and none the projects I’ve worked on since joining my lab have made it to print yet.)
Random thought:
-Brachypodium is the first sequenced grass that hasn’t been domesticated as a crop. I would expect at least a couple of papers will eventually be published that capitalize on that distinction.
*The International Brachypodium Initiative, “Genome Sequencing and analysis of the model grass Brachypodium distachyon” DOI: 10.1038/nature08747 The genome sequence itself can be accessed at here among other places.
**Especially on urban and suburban campuses, land for a cornfield represents a substantial investment.
***No growing plants in winter unless you’ve got enough green house space (and even then they may not be happy enough to flower), or the money to run a winter field somewhere warm like Hawaii or Puerto Rico. And no way to fix it in August if you realize you didn’t plant enough plants to do get everything you need done.
****The number of people will break into a building to pull plants out of a growth chamber because they (usually mistakenly) have decided they plants are genetically engineered and need to be kills is much lower than the number who will do the same thing to an unguarded cornfield.

Nice way to start with a Nature paper!
Comment by Noah Fahlgren — February 11, 2010 @ 5:30 pm
Haha thanks. Congratulations to you and your lab mates too.
Comment by James — February 11, 2010 @ 6:44 pm
Now that I think of it, I guess I had heard of people pulling up “genetically engineered” research crops, but I’d forgotten all about it until you mentioned it here. And people wonder why I hate people.
Also, I don’t quite understand what the blue/cyan dotplot signifies (kinda I do, but kinda I don’t), but I find it a fascinating way of presenting the information.
Comment by mr_subjunctive — February 11, 2010 @ 7:25 pm
I don’t think it happens too often, but for grad students growing plants that may have taken two or three years to create (just using normal cross pollination techniques) and are still a year away from the plants they need to actually complete their research, being forced to start over from scratch is devastating. That’s not the sort of research I do anymore, but you should see the security we have to go through just to get into the Berkeley corn field.
I know didn’t go into any real depth explaining to dotplot, so I’m glad it was still of interest. I think it’s a very visually appealing way of presenting information about a genome which is why I wanted to put it in.
Comment by James — February 12, 2010 @ 8:39 am
There are only 2 academic labs in all of France that still work with transgenic crop species because the destruction of experiments has been so aggressive (and apparently no one has gone to jail, so crime without consequence!). Even these labs send a lot of experimental plants to collaborators in the US to be grown more safely.
Comment by Amy — February 14, 2010 @ 12:35 am
Jeez. I had no idea it had gotten so bad in France (and I’d assume similarly other parts of Europe.)
Comment by James — February 14, 2010 @ 10:08 am
congrats, James
Comment by Haibao Tang — February 11, 2010 @ 7:56 pm
Thanks!
Comment by James — February 12, 2010 @ 8:39 am
Exciting…
Comment by Aron — February 12, 2010 @ 6:13 pm
Brachypodium distachion
Comment by Neji — February 7, 2011 @ 5:35 am