Here’s the key statistic: The maize genome paper estimated that roughly a quarter of maize genes are currently retained as duplicate pairs from maize’s whole genome duplication, while the soybean paper estimates just over half of soybean genes are similarly retained after soybean’s (apparently slightly older) duplication. <– had it buried at the end of this, but figured it’d be more fun to start out with something cool.
But first of all, let’s do this the right way this time. Here’s the paper in Nature describing the soybean genome. Here’s one of the places you can download the entire sequence from. Hopefully that establishes, beyond a reasonable doubt, that the soybean genome has, in fact, been published.
The soybean genome has doubled twice since its last common ancestor with other published genomes like grape and arabidopsis. The genome paper dates the more recent duplication as ~13 million years old and the older as ~59 million years old. When I look at homeologous* regions in soybean from the most recent genome duplication, they look more conserved than comparable regions in corn which also underwent a whole genome duplication relatively recently (the maize genome paper estimated the date at 5-12 million years ago), when one would expect a slightly older duplication to be either less conserved, or at most indistigushable from the younger. Dating genome duplications is difficult to do precisely, it’s possible the maize duplication is older and the soybean younger than current measurements suggest. It’s also possible the difference is exaggerated because comparing homeologous maize chromosomes runs into issues with the waves of transposons that have washed over the maize genome in the past ten million years. The favorite explanation I’ve thought of so far is that the different selective pressures and/or reproductive strategies of the wild ancestors of corn and soybean favored or permitted higher loss of duplicate genes in maize or selected for the avoidance of any gene loss in soybean. No idea how to test any of these ideas though…
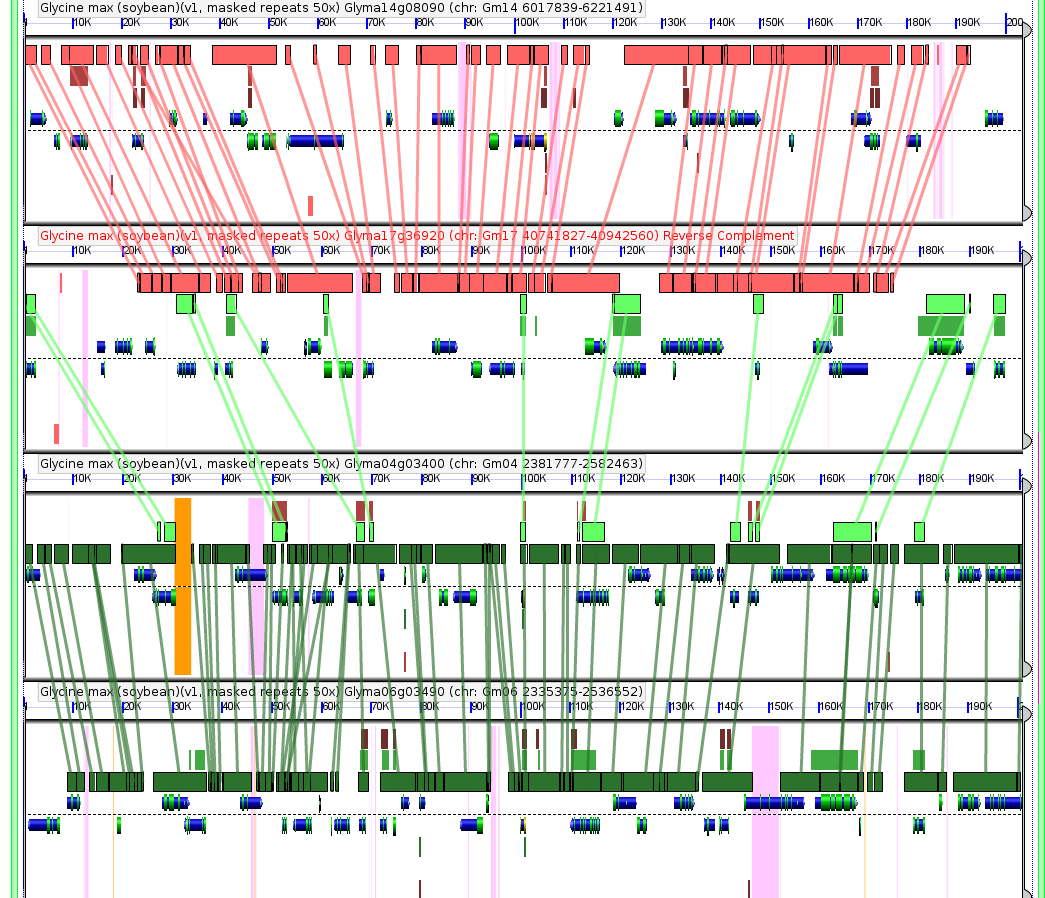
Comparison of four homologous regions in soybean resulting from the two tetraploidies described in the soybean genome paper. Top two and bottom two are related by the recent tetraploidy, and the top and bottom pairs are related by the older tetraploidy. To load this figure live in CoGe (where you can examine the quality of the blast hits, the identity of the genes and generally have fun) visit: http://tinyurl.com/ydh95oh
After a whole genome duplication, every single gene exists as two separate copes on two different chromosomes. Generally speaking two things can happen to a pair of genes. One possibility is that both copies will be retained for either dosage reasons** or because one copy will take on a new function a change called neofunctionalization, or because each of the new gene copies has specialized in part of the job of the original gene, a change called subfunctionalization. The other, generally more common fate of duplicated genes is that one disappears from the genome. Plant genomes are dynamic places (especially compared to animal genomes, but more on that later) and sequences that don’t provide some benefit aren’t preserved for long***. A lot of my own research has to do with how a genome sheds duplicate genes, and the biases in which kinds of genes are lost and which are preserved and I find it fascinating stuff.
Anyway, to all those involved in sequencing the soybean genome, thank you for giving me even more fascinating information to study.
Previous in depth post on soybeans.
*Homeologous regions are areas of the genome that started out as duplicate copies of the same part of the genome after a whole genome duplication, but have since been evolving on their own, usually in different directions.
**If one set of genes produces a protein that works together with many other genes, doubling all of them at once has no stong effect, but if only one gene loses its extra copy, the ratios of the different proteins could end up out of balance, harming the plant. Since it’s highly unlikely that one copy of all the genes involved would be deleted at the same time, the result is that deletions or serious mutations of any of the copies of any of the genes get selected against, and the plant keeps two copies of all the genes involved for longer than other types of genes.
Think of the genome as a restaurant where there’s one person for every job (we’ll simplify it down to just a hostess, and waitress and a cook). Though it wouldn’t in the real work, hypothetically this ratio works out perfectly, the hostess can seat people fast enough to keep the waitress busy but not overwelm her and the cook is constantly cooking but orders aren’t finished late.. If suddenly the owners decide to hire another person for every job so now there are two cooks, two hostesses, and two waiteresses, everything stays in balance. Twice as many people are seated, which is just enough to be handled by two waitresses, and two cooks exactly handle the doubling of orders. But if any one of our six hypothetical people calls in sick the system breaks down. If the hostess calls in sick not enough people are seated and the waitresses, and cooks spend a lot of time just standing around. If one of the waitresses is out, the one waitress has to try to deal with twice as many constomers as she can actually handle, customers aren’t happy because of the poor quality service, and the cooks and dishwashers are still underutilized. If one of the cooks doesn’t show up, people are seated and their orders get taken promptly, but food arrives late, the cook is stressed out, and the customers are unhappy again. If a cook, a waitress, and a hostess all didn’t show up on the same day, everything would be still be in balance, but since sick days (like knock out mutations of genes) occure independently of each other, anyone not showing up to work will be bad for the restuarant, and losing a gene that functions in a complex will be bad for a plant.
***Before anyone points out transposons as a counterexample of sequences that last a long time without, usually, providing a benefit to a plant, keep in mind that because transposons are self replicating, they generate lots of copies. Most of those copies don’t last for millions of years, and almost all those that do are mangled enough by point mutations and insertion deletions that they can no longer function. Similarly when scaning through the genome it’s common to spot pseudogenes. Bits of sequence that used to be a gene, but have been destroyed by