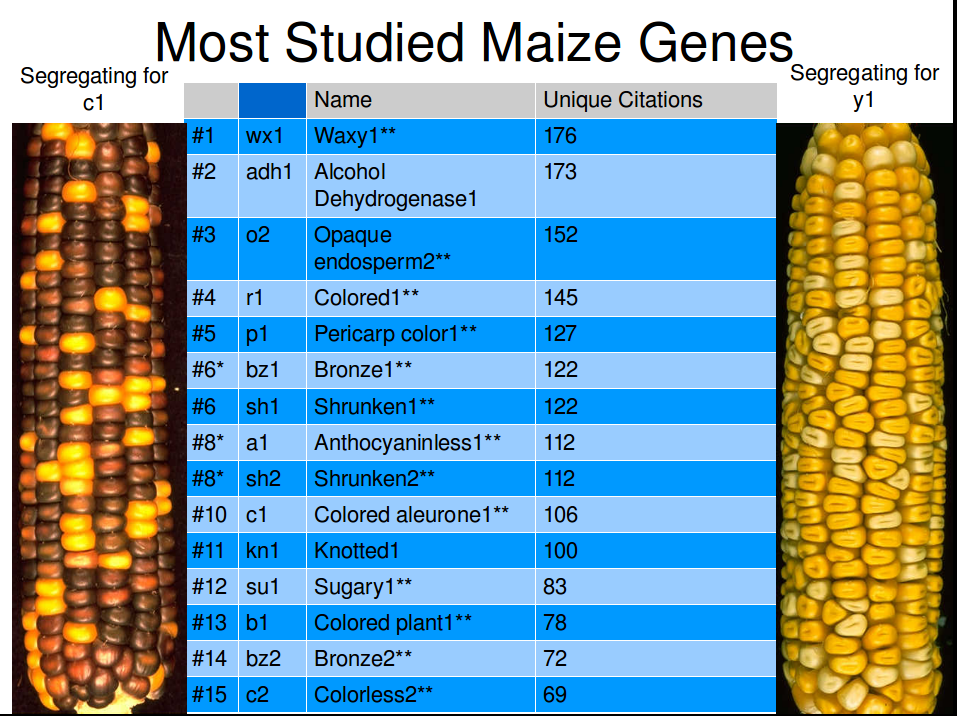
Unique citations determined from papered linked to from MaizeGDB gene locus pages. Images of c1 and y1 segregating years by Gerald Neuffer and made available through MaizeGDB.
* = tied for number of citations
** = some mutant alleles have kernel phenotypes.
If you want to become one of the famous mutant corn genes, it helps if you have an effect that is visible in corn kernels instead of only from fully grown plants.
And here is why:
- A geneticist could determine that the version of c1 that creates yellow kernels is recessive to the version that creates purple kernels just from looking at the ear of corn on left.
- Furthermore, they could tell you that both the male parent (the plant that provided the pollen) and the female parent (the plant on which the ear of corn grew) were both heteryzygous for the c1 genes (they each had one dominant version of the genes and one recessive version), and therefore the corn kernels the parent plants were grown from were both purple.
- They would know with certainty that all of the yellow kernels contain two recessive versions of the c1 gene.
- While they couldn’t predict with absolute certainty whether a specific purple corn kernel on that ear carried two dominant versions of the c1 gene or one dominant and one recessive version, they would know there was a 1/3 chance that kernel has two dominant copies, and a 2/3 chance it had one dominant and one recessive copy.
- That geneticist could make all sorts of predictions about what ears would look like in future generations depending on what colors of corn kernels were planted and which plants were mated with each other.
All this from a single picture of an ear of corn. For a phenotype seen in corn plants but not in kernels (like Knotted1), a geneticist would have to plant a row or more of corn seeds from an ear and examine the growing plants to get the same quantity of information.
And that is why mutations with kernel phenotypes have been so popular over a century of maize genetics research.