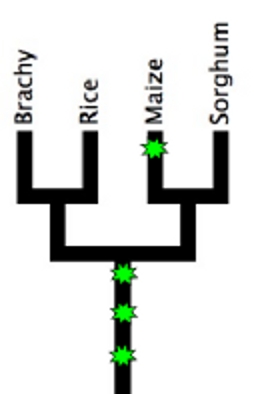
There is a piece of DNA that is sometimes found on the end of the tenth maize chromosome. In plants that possess this extra chromosome segment, chromosome knobs* (including one that’s a part of the extra segment included in abnormal chromosome 10) start to act like centromeres**. But this story graduates from odd to downright weird when I tell you that possessing this extra centromere-like activity gives a chromosome an unfair advantage in being passed on to the next generation.
Plants, like animals, possess two complete genome copies, one from each parent. They’ll only pass on one copy (mixtures of pieces from each parent) to their offspring. Any given sequence has a 50% chance of being passed on which seems fair given the plant is passing on 50% of its total genetic material. But abnormal chromosome ten cheats (using those extra centromere-like sequences I mentioned earlier). It has up to an 83% chance of being passed on.
Since the breed of corn (B73) the maize genome was based on has the normal version of chromosome 10, we know very little about the extra DNA found in abnormal chromosome 10. The authors of this poster are going to correct that oversight, by sequencing the region, figuring out how (and how long ago) abnormal chromosome 10 came into being, and hopefully identifying the genes within the region that make chromosome-knobs act like centromeres.
 *Knobs are dense segments of DNA that scientists have been able to spot visually within chromosomes since before we knew for sure that chromosomes carried genetic information.
*Knobs are dense segments of DNA that scientists have been able to spot visually within chromosomes since before we knew for sure that chromosomes carried genetic information.
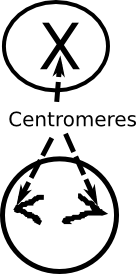
**Centromeres are the part of the chromosomes that bind together during cell division (the center of the X in the traditional drawing of a chromosome). They’re also the place where the molecular machinery that pulls chromosomes apart at the end of the process of cell division.
Lisa Kanizay and Kelly R. Dawe “Uncovering the sequence and structure of maize abnormal chromosome 10” Poster #165 2010 Maize Meeting.